Introduction to Python Programming
Overview
Teaching: 30 min
Exercises: 30 minQuestions
How Python in used in Scientific Computing?
Objectives
Learn the different context on which Python is used.
Python is an interpreted language that is relatively easy to learn and very suitable for scientific computing. The only drawback is performance, but with appropriated arrangements this can be overcome in most cases.
Where to use Python?
From the scientific computing point of view, Python is good to be used in a variety of contexts:
-
Interactive Computing: Using IPython, you can use an interactive shell that allow you to write and execute commands “on the fly”, like you typically do on a calculator. This is often what you do when you are exploring a new library, doing some tests or trying to produce plots for a publication. IPython offers some niceties, like executing shell commands directly from the same IPython shell, read documentation for other python modules and its internal structure (methods and classes) and stored all the commands executed into log files that can be converted into scripts later on.
-
Scripts: Scripts are usually single documents pieces of code that execute some operation, create some figures or do some computation in a rather straightforward way. They are intended to do their job and no more than that. Usually they are short, dirty in the sense that are probably not design to cover all possible error conditions. Scripts are created with a very pragmatic view.
-
Executable Programs: Like an evolution of scripts, an full-featured program executes like a command line program, it probably started as an script and evolve by having several functions collected in modules, they grew in complexity to the point where you can start calling it from the command line with arguments that are captured by the program and works with then to produce a result, that being a report file, a table, a figure or something else.
-
Libraries: Libraries are collections of functions or even more complex code structures, that are intended to be used by another Executable Program, or script or being used during an interactive session. The fact that they are not linked to a defined execution pathway. Libraries are created as general as possible. As the developer using the library could be different from who developed the library, documentation is crucial to make the library useful to others.
-
GUI Applications: Graphical User Interface (GUI) applications are another way of creating applications where the final user interacts directly with the keyboard and mouse and the application is displayed as a windows on the users computer.
-
Web Applications: Web applications are very special case of GUI where the window is replaced by a browser tab and the interaction usually happens with a remote application on the web. Python offers packages for this kind of use case, like Flask and Python bottle up to full featured solutions like CherryPy and Django.
This taxonomy is exempt of inconsistencies and there are cases where software packages fits under several of those categories. What is important to realize is that Python is a very flexible language suitable to offer solutions under a variety of conditions.
Organization of this Workshop
This is 2-day workshop, very practical with examples and exercises to provide exposure to the concepts presented during the episodes. The first day will be focused on the Python language itself and the Standard Library, a set of modules that are always present with any python deployment. We will work entirely with Python 3.6 or Python 3.7, the old Python 2.7 will retire and will not be maintained past 2020 even if there is a bunch of code written in Python 2.x.
The second day is focused on scientific computing with Python:
-
Numpy: The key component for most you can do with python in terms of numerical programming. Numpy offers an interface between python and the linear algebra routines BLAS and LAPACK that are critical on many numerical computing operations.
-
Scipy: A more elaborated library of numerical routines including special functions, integration and interpolation among several others.
-
Matplolib: The most commonly used plotting package in Python.
-
Pandas: Provides data structures for data analysis, including the ability to read data from several formats and dealing with missing and wrong data.
-
Cython Cython is a library that converts python code into C in order to optimize the execution of computational expensive operations without having to loose completely the flexibility that python provides.
Getting Python up and Running
If you are working on Thorny Flat, all that you have to do is loading the environment module that will give you access to Python 3. In general HPC clusters today include old versions of python and usually limited to Python 2.x
module load lang/gcc/8.2.0 lang/python/3.7.2_gcc82
or
module load lang/python/intelpython_3.6.3
Both options should give you access to a recent version of Python 3 with a number of scientific packages that we will use for the second day.
Assuming that you load the environment module for CPython, the original version of Python implemented in C, you can check the version of the interpreter with:
python3 --version
Python 3.7.2
Notice that the command python could still point to the 2.x version, so always use python3 to ensure you are using the right version.
Python comes with a interactive interpreter, we will not use it here but it is good for you to know that just executing python3 will give you the standard python interpreter.
$ python3
Python 3.7.2 (default, Feb 12 2019, 17:15:23)
[GCC 8.2.0] on linux
Type "help", "copyright", "credits" or "license" for more information.
>>>
Try simple calculator-like commands, for example 2+2, to leave the interpreter, execute exit() or the combination of Control (CTRL) and the key D, we express this kind of combination as CTRL-D
This interactive interpreter is sufficient for small executions, but there is a much better replacement including a wealth of features much better suited for scientific exploration, it is called IPython and it can be accessed with:
$ ipython3
Python 3.7.2 (default, Feb 12 2019, 17:15:23)
Type 'copyright', 'credits' or 'license' for more information
IPython 7.5.0 -- An enhanced Interactive Python. Type '?' for help.
In [1]:
This interpreter offers a far more pleasing experience, allowing you to go back to previous commands, recovering the output of previous executions, storing the history of commands in logs and exploring the documentation of other python modules. We will use IPython for most of our exploration of the basics of the python language.
Testing Python with X11 Forwarding
To test if you are able to get remote windows popping on your desktop machine. Two conditions have to be ensured:
-
You have a XServer running on your machine. If you are using MacOS you have to install XQartz https://www.xquartz.org/, if you are using Windows, consider installing a SSH client with Xserver incorporated such as MobaXterm https://mobaxterm.mobatek.net/. If you are using Linux, you do not need to do anything special.
-
You connect to our cluster with X11 port forwarding. If you are using Linux, or MacOS or MobaXterm on Windows all that you have to do is execute:
$ ssh -Y -C username@spruce.hpc.wvu.edu
Once you have a SSH connection to our cluster. Test that you get a simple window back to your desktop
$ xeyes
or
$ xclock
Now, lets load one of modules with Python 3, for example using CPython will be:
$ module load lang/python/3.7.2_gcc82
Execute IPython:
$ ipython3
Python 3.7.2 (default, Jan 11 2019, 13:58:19)
Type 'copyright', 'credits' or 'license' for more information
IPython 7.4.0 -- An enhanced Interactive Python. Type '?' for help.
In [1]:
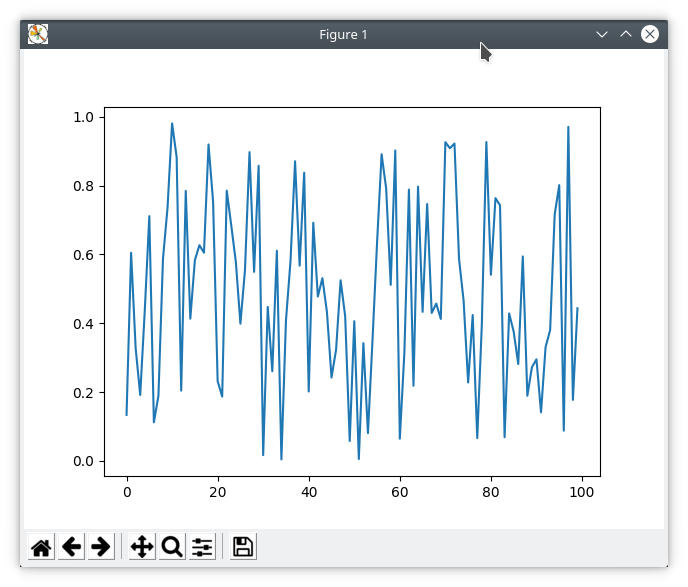
Execute these commands and a plot should appear on your computer.
$ ipython3
Python 3.7.2 (default, Jan 11 2019, 13:58:19)
Type 'copyright', 'credits' or 'license' for more information
IPython 7.4.0 -- An enhanced Interactive Python. Type '?' for help.
In [1]: %matplotlib tk
In [2]: import numpy as np
In [3]: import matplotlib.pyplot as plt
In [4]: plt.plot(np.random.rand(100))
Out[4]: [<matplotlib.lines.Line2D at 0x7f2f819e8978>]
Using IPython with a Singularity container
There are more ways of using IPython for interactive computing. They require also X11 forwarding.
Connect to the cluster with X11 Forwarding:
$ ssh -Y -C username@spruce.hpc.wvu.edu
Load the module for Singularity containers
$ module load singularity/2.5.2
Enter in a shell session on a container that includes the IPython notebook
$ singularity shell /shared/software/containers/jupyter-xenial.simg
Singularity: Invoking an interactive shell within container...
Singularity jupyter-xenial.simg:~>
For executing the qtconsole:
$ ipython3 qtconsole
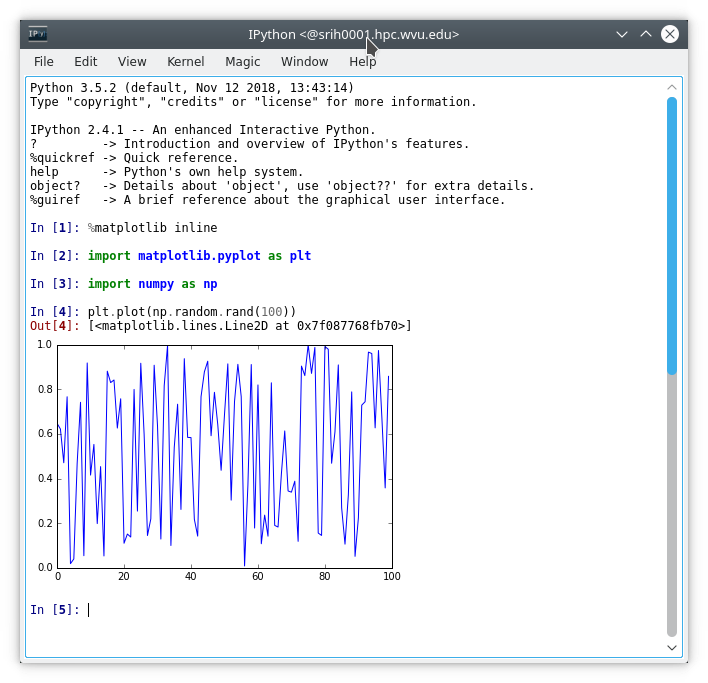
You should see a terminal popping on your desktop and execute the commands:
In [1]: %matplotlib inline
In [2]: import numpy as np
In [3]: import matplotlib.pyplot as plt
In [4]: plt.plot(np.random.rand(100))
Out[4]: [<matplotlib.lines.Line2D at 0x7f2f819e8978>]
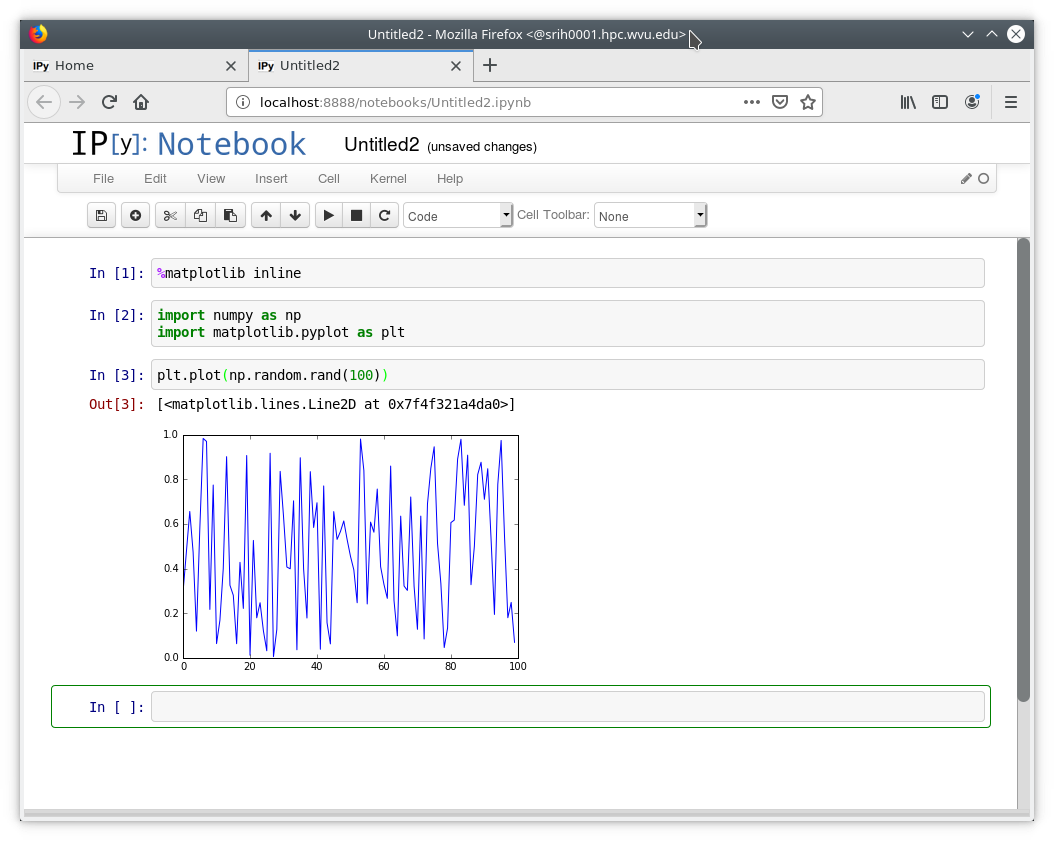
For the notebook
$ ipython3 notebook
Execute the same commands as for the qtconsole. In order to execute each command enter Shift-ENTER
Key Points
Understand the difference between interactive computing, scripts and programs.
Checking the version of Python that is working on the system.